Immunoinformatics prediction for designing potential epitope-based malaria vaccine in the Aminopeptidase N1 protein of Anopheles gambiae (Diptera: Culicidae)
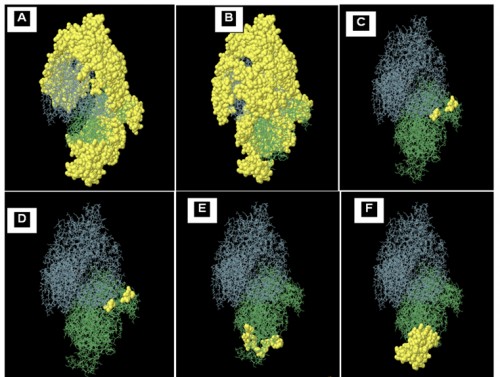 Fig. 1:
Fig. 1: 2D view of conformational epitopes on four chains (A to D) of APN 1 protein. The epitopes showed by yellow surface, and the bulk of the grey and green sticks represents chain A and chain B, respectively of APN1 protein. E and F represent the surface Accessibility of VDERYRL and VQYSTDT, B cell epitopes respectively on B chain.
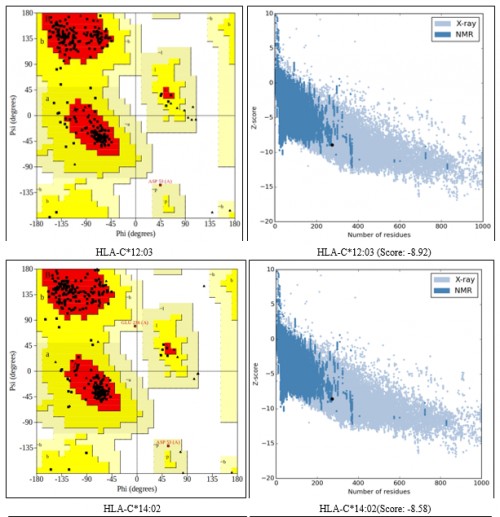 Fig. 2:
Fig. 2: (A) HLA-C*12:03, (B) HLA-C*12:03 (Score: -8.92), (C) HLA-C*14:02, (D) HLA-C*14:02(Score: -8.58); Ramachandran Plot and Prosa Z-score validation of HLA-C*12:03, HLA-C*14:02 and HLA-A*32:01.
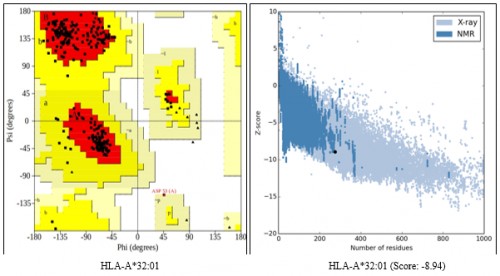 Fig. 3:
Fig. 3: (A) HLA-A*32:01, (B) HLA-A*32:01 (Score: -8.94) Ramachandran Plot and Prosa Z-score validation of HLA-C*12:03, HLA-C*14:02 and HLA-A*32:01.
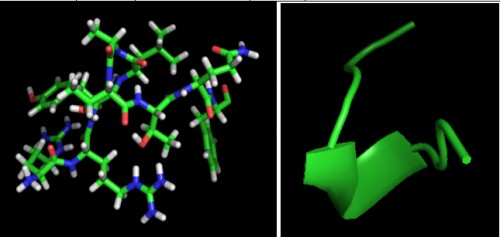 Fig. 4:
Fig. 4: Peptide Structure of predicted epitope “RRYLATTQF†for MHC I prepared by PEP-FOLD.
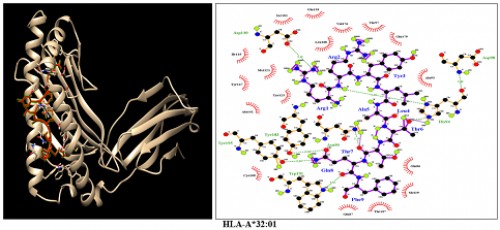 Fig. 5:
Fig. 5: HLA-A*32:01;Docking sites of predicted peptide (RRYLATTQF) against selected MHCI receptors.
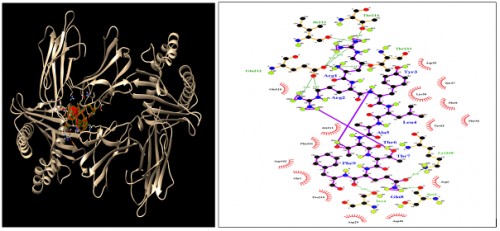 Fig. 6:
Fig. 6: HLA-B*27:05; Docking sites of predicted peptide (RRYLATTQF) against selected MHCI receptors.
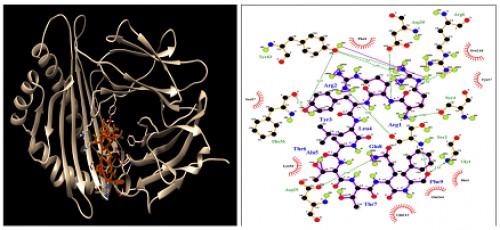 Fig. 7:
Fig. 7: HLA-C*03:03; Docking sites of predicted peptide (RRYLATTQF) against selected MHCI receptors.
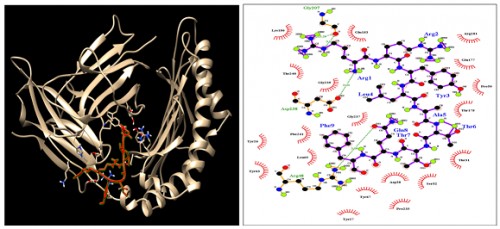 Fig. 8:
Fig. 8: HLA-C*07:02; Docking sites of predicted peptide (RRYLATTQF) against selected MHCI receptors.
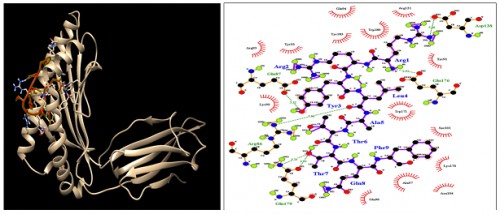 Fig. 9:
Fig. 9: HLA-C*12:03; Docking sites of predicted peptide (RRYLATTQF) against selected MHCI receptors.
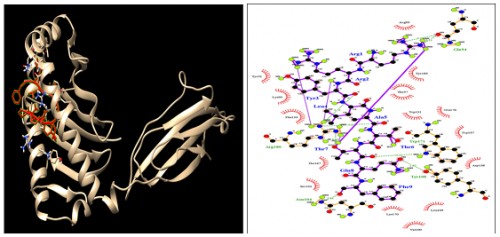 Fig. 10:
Fig. 10: HLA-C*14:02; Docking sites of predicted peptide (RRYLATTQF) against selected MHCI receptors.











