Genetic diversity studies on Culex quinquefasciatus from diverse larval habitats using RAPD-PCR marker
Author(s): Shabad Preet and Shivani Gupta
Abstract: The study focuses on the analysis of population genetic structure of Culex quinquefasciatus from various larval habitats using RAPD- PCR. The population differentiation parameters, genetic distances, molecular variance and cluster analysis were done using POPGENE, GenALEx and MEGA softwares. Out of 140 RAPD primers screened, four primers exhibited distinct RAPD banding patterns showing up to 100% polymorphism. Moderate variation was evident from value of GST among larval populations. Effective migration rates were observed to be depicting high gene flow. The AMOVA analysis revealed high intrapopulation genetic variation (92%) with only 8% variation among populations. The cluster analysis showed one main cluster which is divided into two subclusters, one between pond and waste water populations and another cluster of cement tanks along with plastics as a separate branch. The study revealed establishment of Cx. quinquefasciatus in diverse larval habitats with excellent adaptability as indicated by high gene flow and polymorphism.
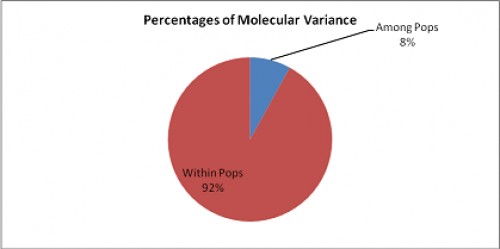 Fig.:
Fig.: Percentages of molecular variance in
Cx. Quinquefasciatus populations
How to cite this article:
Shabad Preet, Shivani Gupta. Genetic diversity studies on Culex quinquefasciatus from diverse larval habitats using RAPD-PCR marker. Int J Mosq Res 2017;4(2):93-98.



