Engineering lhRNA based molecule to interfere replication of dengue virus in transgenic Aedes aegypti mosquitoes: Bioinformatics approach
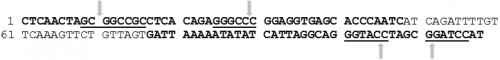 Fig. 1:
Fig. 1: Ae. aegypti sialokinin
I intron sequence (5’ to 3’). Forward and reverse primers are shown in bold black letters. The underlined sequences are restriction recognition sites (REs),
Not
I, Apa
I, Kpn
I and BamH
I respectively from the 5’ end of the sequence. Each arrow indicates the cleavage site within the
REs. Fig. 2:
Fig. 2: Mnp+ (A) regions (5’ to 3’) selected from SLDENV-2 (gb: GQ252677). Mnp+ and Mnp- forward and reverse primers are shown in bold black letters. The underlined sequences are REs,
NotI and
ApaI respectively from the 5’ end of the Mnp+ sequence and
KpnI and
BamHI respectively from the 5’ end of the Mnp- sequence. These sequences had a 100% identity with another SLDENV-2 strain (gb: GQ252676). But there were a few mismatches in the sequence when these two sequences were aligned with that of the Mnp+ region of the SLDENV-2 strain found in 1996 (gb: FJ882602). The sites of sequence variations, base 54th position (A), 94th position (A), 109th position (C), 157th position (T) and 211th position (T) are shown in bold black letters between the primer sequences of Mnp+ sequence. Each arrow indicates the cleavage site within the REs
 Fig. 3:
Fig. 3: Mnp- (B) regions (5’ to 3’) selected from SLDENV-2 (gb: GQ252677). Mnp+ and Mnp- forward and reverse primers are shown in bold black letters. The underlined sequences are REs,
NotI and em>ApaI respectively from the 5’ end of the Mnp+ sequence and
KpnI and
BamHI respectively from the 5’ end of the Mnp- sequence. These sequences had a 100% identity with another SLDENV-2 strain (gb: GQ252676). But there were a few mismatches in the sequence when these two sequences were aligned with that of the Mnp+ region of the SLDENV-2 strain found in 1996 (gb: FJ882602). The sites of sequence variations, base 54th position (A), 94th position (A), 109th position (C), 157th position (T) and 211th position (T) are shown in bold black letters between the primer sequences of Mnp+ sequence. Each arrow indicates the cleavage site within the REs
 Fig. 4:
Fig. 4: The multiple sequence alignment showing the transcription initiation site (+1) and the translation initiation site of the AeCPA gene.
The alignment of whole genome sequence of
Ae. aegypti, gb: AAGE02019343.1 (A), complete cDNA sequence of AeCPA gene, gb: AF165923.1 (B), final transcript of AeCPA gene, gb: AF165923.1 (C), and the reverse translated amino acid sequence, gb: AAD47827.1 (D) of the AeCPA is shown. The numerical values at the top of the figure show the position of each nucleotide in the sequence, AAGE02019343.1. Hence, it can be seen that the 1442th position, of genome sequence, AAGE02019343.1 of AeCPA gene is the transcription initiation site of the gene and the 2132th position of the sequence is the translation initiation site
 Fig. 5: Ae. aegypti
Fig. 5: Ae. aegypti Carboxypeptidase A promoter sequence from -4 to -1126 (5’ to 3’). The
AeCPAF and AeCPAR primers are shown in bold black letters. The underlined sequences are REs,
SacI and
SmaI respectively from the 5’ end of the sequence. Each arrow indicates the cleavage site within the REs
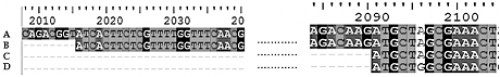 Fig. 6:
Fig. 6: The multiple sequence alignment showing the transcription initiation site (+1) and the translation initiation site of the AeVtA gene.
The alignment of complete DNA sequence of
Ae. aegypti VgA1 gene, gb: L41842.1 (A), the complete cDNA sequence of the very sequence (B), the final transcript of the very sequence (C), and the reverse translated amino acid sequence, gb: AAA99486.1 (D) is shown. The numerical values at the top of the figure show the position of each nucleotide in the sequence, gb: L41842.1. Hence, it can be seen that the 2016th position, of gene sequence, gb: L41842.1 of AeVtA gene is the transcription initiation site of the gene and the 2091st position of the sequence is the translation initiation site
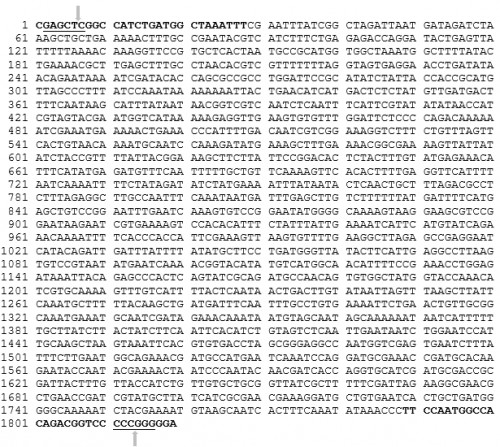 Fig. 7: Ae. aegypti
Fig. 7: Ae. aegypti Vitellogenin promoter sequence
(5’ to 3’). The
AeVtAF and AeVtAR primers are shown in bold black letters. The underlined sequences are REs, SacI and SmaI respectively from the 5’ end of the sequence. Each arrow indicates the cleavage site within the REs.
 Fig. 8:
Fig. 8: svA sequence (5’ to 3’).
svA forward and reverse primers are shown in bold black letters. svA region of both vectors, pGL3-hsp82 (Addgene) and pGL3-Basic (Addgene) could be amplified by the primers indicated. The underlined sequences are REs,
BamHI and
XbaI respectively from the 5’ end of the sequence. Each arrow indicates the cleavage site within the REs.
 Fig. 9:
Fig. 9: MosI transposase enzyme coding sequence.
The MosF and MosR primers are shown in bold black letters. The underlined sequences are REs,
NcoI and
FseI respectively from the 5’ end of the sequence. Each arrow indicates the cleavage site within the REs.
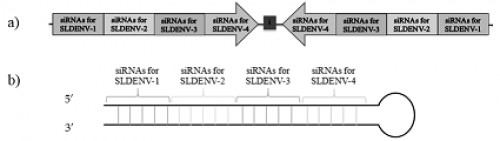 Fig. 10:
Fig. 10: The designed effector lhRNA molecule in this study. The construct showing different regions of lhRNA made up of siRNAs specific to different dengue virus serotypes (a) The stem and loop structure of lhRNA molecule formed by complementary base paring of its sense and antisense regions (b). The sequence of designed lhRNA molecule that corresponds to positive (+) strand of the stem structure, the position 799 to 874 is the region that corresponds to loop structure and the position 874 to 1672 is the region that corresponds to negative (-) strand sequence of the stem structure of lhRNA molecule. The spaces in the figure indicated by two arrows (at the top of the figure) are the sites of lhRNA molecule that are cleaved by the dicer enzyme and the regions between spacer are the designed siRNA sequences containing 21 nucleotides.
 Fig. 11:
Fig. 11: The designed effector lhRNA molecule in this study. The construct showing different regions of lhRNA made up of siRNAs specific to different dengue virus serotypes (C), and the position 1 to 798-nt of the sequence is the region that corresponds to positive (+) strand of the stem structure, the position 799 to 874 is the region that corresponds to loop structure and the position 874 to 1672 is the region that corresponds to negative (-) strand sequence of the stem structure of lhRNA molecule. The spaces in the figure indicated by two arrows (at the top of the figure) are the sites of lhRNA molecule that are cleaved by the dicer enzyme and the regions between spacer are the designed siRNA sequences containing 21 nucleotides.












